This tutorial will take you through the same steps I took when I was asked to see the feasibility of a lightweight fingerprinting system based on Gaia.
I was actually surprised to have something up and running very quickly, so I thought that the time I spared doing this, I would share it with others to teach them how to use Gaia. (Another reason, more tangible, is that my computer is so slow currently due to itunes db untar'ing and merging that I cannot do anything useful except documentation...)
NB: Of course, the "less than an hour" must have told you we were going to use the python bindings, so you will need that. I strongly suggest to use IPython instead of the standard python interpreter if you are going to to interactive stuff, because it is so much better: colors, autocompletion, integration with pylab so you can have multiple plots at the same time, etc...
Setting up the required DataSet, etc...
NB: This part is done in a command-line shell, and not yet in the python interpreter.
The first thing we will need to do, obviously, is to get a database. I will assume you have run Essentia on your files, and that they are stored in a directory, let's call it descriptors/, for instance (rings a bell?) :-)
If you don't want to come up with specific IDs for each file, there is a very convenient script, located at gaia/src/scripts/make_db_from_directory, which will recurse a directory, find all .sig files in there, and write a yaml file containing the name of the file (without '.sig') as the key, and its full path as the value. Which is exactly what you need to merge all your Essentia files into a Gaia DataSet. This gives:
$ ~/dev/gaia/src/scripts/make_db_from_directory.py fingerprint descriptors/
This will create a file named fingerprint.dbfiles.yaml in the directory you are currently in. Let's merge the database now.
$ gaiamerge fingerprint.dbfiles.yaml fingerprint.db
You now have the DataSet ready to be processed! Great!
Processing to DataSet to have it in a better shape (more suited to fingerprinting tasks)
NB: You should now be in a python shell (preferably IPython)
Now that we have our DataSet ready, we want to process it. Here, as we want to do fingerprinting, we need something that is not necessarily musically precise, but it should be as fast as possible. For these reasons, we will clean/normalize as usual, and then apply a PCA with a low target dimension, such as 10 or 15, for instance. Let's do that:
from gaia2 import *
# first load the dataset
ds = DataSet()
ds.load('fingerprint.db')
# let's transform it so it is in a nice shape for us
ds_novl = transform(ds, 'removevl')
dsc = transform(ds_novl, 'cleaner')
dsn = transform(dsc, 'normalize')
dspca = transform(dsn, 'pca', { 'dimension': 15 })
Finding a good threshold for identifying duplicates
Here we go! Now, the idea we want to exploit here, is that if there are duplicates, their distance to the query point should be very small (ideally 0). So we need to set a threshold for the distance under which the points are going to be considered duplicates. However this threshold will highly depend on the descriptors computed by essentia, the type of normalization that was used, the final number of dimensions in the PCA, etc...
What we will do here then, is for each point of the dataset, get its 10 nearest neighbours, and plot the histogram of the distances we get.
# prepare distance, view
euclid_dist = MetricFactory.create('euclidean', dspca.layout())
v = View(dspca, euclid_dist)
# ...and go for it!
distances = []
for point_query in dspca.points():
results = v.nnSearch(point_query).get(10)
for (p, dist) in results:
distances.append(dist)
# now we plot it
import pylab
pylab.hist(distances, bins=100)
pylab.show() # not necessary if you started IPython with the -pylab option
This gave me the following graph (YMMV):
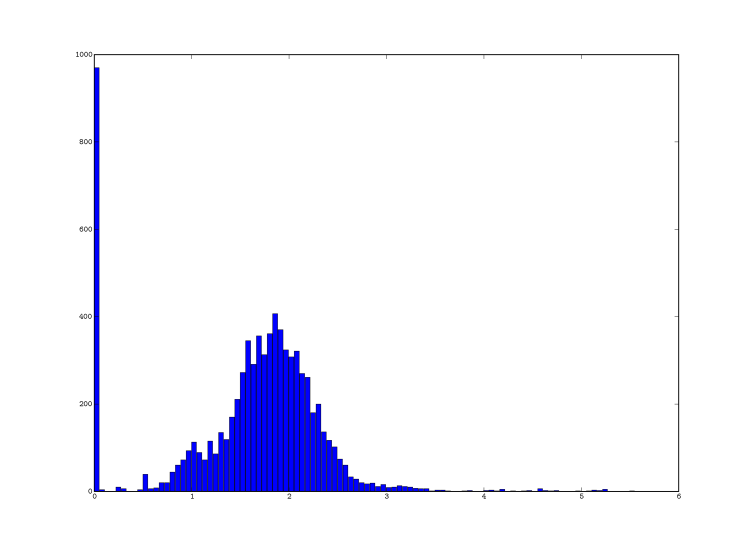
Based on this, I decided to set the threshold for fingerprinting to 0.4 (which is just before the small peak).
Final step: get the list of duplicates
So now, back to finding our duplicates: we want to know for each point, which are its neighbours which have a distance which is less than 0.4:
duplicates = {}
for point_query in dspca.points():
results = v.nnSearch(point_query).get(10)
query_id = point_query.name()
doubles[query_id] = []
for (p, dist) in results:
if p != query_id and dist < 0.4:
doubles[point].append(p)
# that's it, let's print the results
print 'duplicates list:'
for p in double.keys():
# we only want to know which files have duplicates, hence the check
if doubles[p]:
print p, ':', doubles[p]
And that's it! You have the list of duplicates in your database. Of course, you will probably need to evaluate these results, adjust the threshold, etc... But I'll leave this up to you!
